Research
I Application of DNA markers in Basmati rice
Rice is the staple food for more than half of the world's population. In the evolution of rice and its genetic differentiation into distinct varietal groups, consumer quality preferences have played a significant role besides agro-ecological factors. One such varietal group comprising of aromatic pulao/biryani rice of Indian sub-continent known as 'Basmati' is the highly priced rice in the domestic as well as international markets. Originated in the foothills of the Himalayas Basmati rice is characterized by extra long slender grain, pleasant and distinct aroma and soft and fluffy texture of cooked rice. These unique features of Basmati said to be the culmination of centuries of selection and cultivation by farmers, are well preserved and maintained in their purest form in the traditional Basmati (TB) varieties. The historical and archeological findings infer that the varieties with such unique morphological and quality attributes are not present in traditional rice growing areas anywhere in the world. TB rice is not only in great demand in the domestic markets, but is also seen in the menu of connoisseurs worldwide. Basmati rice qualifies as a Geographical Indication (GI).
Keeping the foregoing in view studies were undertaken with the following objectives:
- Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties
- Detection and quantification of adulteration in Basmati rice
1. Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties
Traditionally used morphological and chemical parameters have not been found to be discriminative enough, warranting more precise techniques. Several molecular techniques are available for detecting genetic differences within and among cultivars. Among these, simple sequence repeat (SSR) markers are efficient and cost-effective and detect a significantly higher degree of polymorphism in rice. They are ideal for genetic diversity studies and intensive genetic mapping. An alternative method to SSR, called inter-SSR (ISSR) PCR, has also been used to fingerprint the rice varieties. In the present study, we automated the ISSR-PCR marker assay to enhance genetic informativeness and used it along with rice SSRs to analyze the genetic relationships of TB, EB, and NB varieties.
A subset of three rice groups was analyzed by using 19 simple sequence repeat (SSR) loci and 12 inter-SSR-PCR primers. A total of 70 SSR alleles and 481 inter-SSR-PCR markers were revealed in 24 varieties from the three groups. The lowest genetic diversity was observed among the traditional Basmati varieties, whereas the EB varieties showed the highest genetic diversity by both the marker assays. The results indicated that the subset of aromatic rice varieties analyzed in the present study is probably derived from a single land race. The traditional Basmati (TB) and semi-dwarf NB rice varieties used in the present study were clearly delineated by both marker assays. A number of markers, which could unambiguously distinguish the TB varieties used in the present study from the evolved and NB rice varieties, were identified. The potential use of these markers in Basmati rice-breeding programs and authentication of TB varieties used in the present study are envisaged.
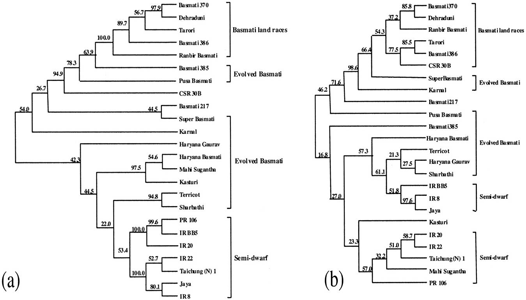
Figure 1: Dendrograms derived from an unweighted pair group method analysis (UPGMA)
cluster analysis by using Nei and Li Coefficients based on (a) ISSR markers and (b)SSR markers.
2. Detection and quantification of adulteration of Basmati rice
Basmati rice, in addition to the desired quality traits, harbors many undesirable traits that include tall stature, low yield and photosensitivity. Attempts since 1970s to develop high yielding Basmati rice varieties by cross breeding, though, resulted in many high yielding Evolved Basmati (EB) varieties; they fell short of the quality standards of TB varieties. The evolved and non-Basmati rices looking similar to TB varieties in appearance have suited the interests of unscrupulous traders to use them as adulterants in Basmati trade. The practice of Basmati adulteration in the long run, would seriously jeopardize our export trade of over Rs.2000 crores. Hence, identifying genuine Basmati variety from other look-alike long grain non-Basmati varieties is important from the viewpoint of sustainable trade. Traditionally employed morphological and biochemical assays for detecting adulteration in Basmati rice have not been found to be discriminative enough warranting more precise high-throughput techniques.
A capillary electrophoresis [CE] based microsatellite DNA profiling protocol was developed, which facilitates quick and accurate detection and quantification of the adulteration in Basmati rice. The single-tube assay multiplexes eight microsatellite loci viz., RM1, RM55, RM44, RM348, RM241, RM202, RM72 and RM171 to generate variety-specific allele profiles that can detect adulteration from 1% upwards (Figures 2 and 3). Accuracy of quantification has been shown to be ±1.5%.
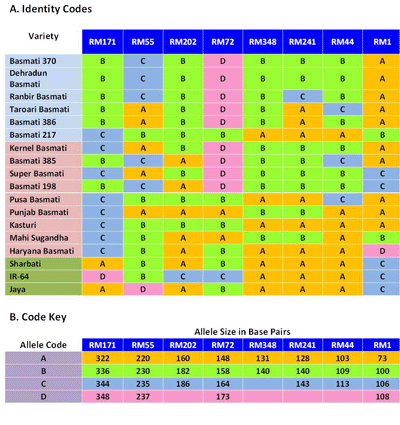
Figure 2: Specific allele profiles of Basmati rice varieties and putative adulterants at eight microsatellite loci.
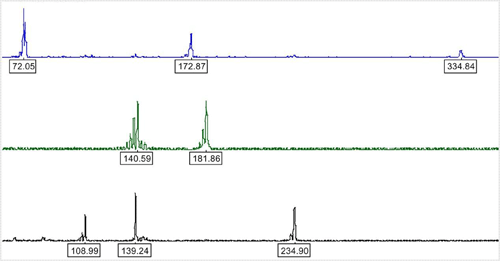
Figure 3: Electropherogram of multiplex allele profile of Dehradun Basmati. Each peak of the multiplex profile represents the variety specific allele at that locus, with allele size annotated in the box. The loci (order: from left to right) are RM1, RM72 and RM171 (in blue); RM241 and RM202 (in green); RM44, RM348 and RM55 (in black).
In addition, it was shown that CE is essential for microsatellite based protocol for detection and quantification of adulteration in Basmati rice by comparing with slab-gel and agarose gel electrophoresis. Comparative analysis across eight microsatellite loci in twelve rice varieties demonstrated that CE method showed less error (±0.73bp) in estimation of allele sizes compared to slab-gel (±1.59bp) and agarose gel (±8.03bp) electrophoretic methods. When the average deviations were further scrutinized for the actual distribution of allele sizes, it was observed that nearly 58% of the alleles estimated by capillary electrophoresis showed exact values (zero deviation). In all, >90% of the capillary electrophoresis assays resulted in allele sizing within a base-pair bin, whereas the corresponding estimate was <50% for slab gel electrophoresis and a dismal 7 and 2%, respectively, for software-based and manually measured agarose gel electrophoresis (Figure 3). Moreover, CE method showed greater reproducibility (<0.5bp deviation) compared to slab-gel (1bp) and agarose (>3bp) based methods. CE method was significantly superior in quantification of the adulterant also, with mean error of ±3.91% in comparison to slab-gel (±6.09%).Lack of accuracy and consistency of the slab-gel and agarose electrophoretic methods warrants the employment of capillary electrophoresis for Basmati rice purity tests.
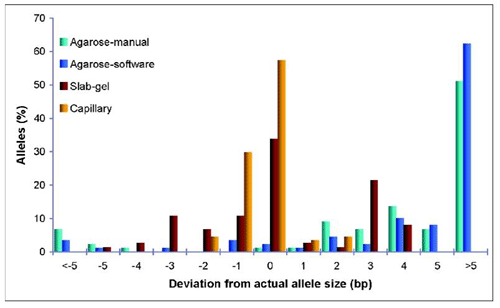
Figure 4: Allele size estimation accuracies of agarose, slab gel, and capillary electrophoresis methods. Y-axis presents fraction of total alleles (expressed in percentage) showing deviations from the actual allele sizes. X-axis shows the deviation (in bp) of the observed allele sizes from the actual allele sizes obtained by bidirectional sequencing. Agarose gel allele sizes were estimated separately by software and manual methods.
II QTL mapping of economically important traits of Basmati rice
Earlier investigations on genetic analysis of different accessions of rice revealed that the Basmati rices form a distinct cluster different from indica, japonica and javanica varietal groups suggesting that Basmati rice falls in phylogenetically divergent group. As for inheritance and breeding behavior are concerned, all the indices of grain appearance and cooking/eating qualities with the exception of amylose content and to some extent aroma have been reported to be polygenically controlled. For instance, amylose content is reported to be governed by single gene located on chromosome 6 i.e., waxy locus (wx) in association with modifier complex. Some researchers are of the opinion, that wx gene also controls gelatinization temperature and gel consistency. In case of aroma, most researchers believe that it is under the control of single recessive gene. Recently, a gene for fragrance (fgr) was identified on chromosome 8 which has homology to the gene that encodes betain aldehyde dehydrogenase 2 (bad2). Thus, the complex genetic architecture of unique traits of Basmati and tedious screening methodologies involved in quality testing have been serious constraints to breeding for quality of Basmati. Moreover, very limited information is available on the genetic basis of grain and cooking qualities of rice in general and basmati rice in particular.
In order to construct Basmati rice linkage map, we crossed a traditional basmati variety Basmati 370 and a semi dwarf variety Jaya to develop a mapping population. 450 microsatellite primers were screened for the polymorphism between parents. Size variation was obtained in 208 loci; polymorphic markers were distributed in all the 12 chromosomes of the rice genome. We obtained an F1 hybrid of the two parental strains, which was confirmed by using two microsatellite markers RM263 and RM339. Phenotypic data of F1 plants were taken. Hybrids exceeded parental strains for all the phenotypic traits. The mapping population has been screened with polymorphic microsatellite markers and constructed a Basmati linkage map and identified many QTLs for different agronomic and quality traits.
Employing F2, F3, and recombinant Inbred Line (RIL) mapping populations derived from the cross between Basmati370 and Jaya, a total of 34 QTLs for 16 economically important traits of Basmati rice were identified. Out of which, 12 QTLs contributing to more than 15% phenotypic variance were identified and considered as major effect QTLs. Four major effect QTLs coincide with the already known genes viz., sd1, GS3, alk1 and fgr governing plant height, grain size, alkali spreading value and aroma QTLs, respectively, For the remaining major QTLs, candidate genes were predicted as auxin response factor for filled grains, soluble starch synthase 3 for chalkiness and VQ domain containing protein for grain breadth and grain weight QTLs, based on the presence of non-synonymous single nucleotide polymorphism (SNPs) that were identified by comparing Basmati genome sequence with that of Nipponbare. To the best of our knowledge, the current study is the first attempt ever made to carry out genome-wide mapping for the dissestion of the genetic basis of economically important traits of Basmati rice. The promising QTLs controlling important traits in Basmati rice, identified in this study, could be used as candidates for future marker-assisted breeding.
|